ParaView
"ParaView is an open-source, multi-platform application designed to visualize data sets of varying sizes from small to very large. The goals of the ParaView project include developing an open-source, multi-platform visualization application that supports distributed computational models to process large data sets. It has an open, flexible, and intuitive user interface. Furthermore, ParaView is built on an extensible architecture based on open standards.
ParaView runs on distributed and shared memory parallel as well as single processor systems and has been successfully tested on Windows, Linux, Mac OS X, IBM Blue Gene, Cray XT3 and various Unix workstations and clusters. Under the hood, ParaView uses the Visualization Toolkit as the data processing and rendering engine and has a user interface written using the Qt cross-platform application framework." link
- ParaView Homepage (http://www.paraview.org)
- ParaView Manuals (http://www.paraview.org/documentation)
- ParaView Prebuild Executables (http://www.paraview.org/download)
- ParaView Wiki (http://www.paraview.org/Wiki/ParaView)
- recommended webpages on more about ParaView
Run ParaView
We recommend to use ParaView on JURECA via VNC:
There are other more advanced configurations that can be used for Remote Visualization or separation of services (Data, Rendering, and User Interface).
Each mode separates the three main components (User Interface, Computation, Rendering) in different ways.
More details on possible ParaView scenarios can be found here or here or here (ParaView on Jureca in VNC session).
Build ParaView
Please check the prebuild ParaView before you build your own ParaView.
If anything is missing in the prebuild ParaView let us know.
You can find ParaView
- as an desktop icon via VNC (profile 'vis').
- on JURECA as a module (
module spider ParaView)
If you want to have your own ParaView you can
- download a prebuild ParaView from the official webpage download.
- build ParaView yourself - check the details of the JURECA build.
Parallel ParaView
If your dataset is large you might be limited by the size of the memory or the cpu performance of a single node.
Executing filters on your data in ParaView might be slow and takes too long or even fails because of memory limitations.
One possible solution can be to run ParaView im parallel across multiple nodes.
How to start ParaView in parallel
First of all, parallel ParaView is only available on vis-compute nodes (not on vis-login nodes).
We recommend the following:
- Start a VNC session on multiple vis-compute nodes using the login tool 'Strudel' (more details).
- Start ParaViews 'pvserver' with MPI multiple times on the nodes
- click the icon 'start PVServers' on the VNC desktop (profile vis).
- or use a script like this after you loaded the ParaView module (srun-pvserver-vis.sh).
- Start ParaView and connect to the pvservers (localhost:11111).
How to use ParaView in parallel
Just starting ParaView in parallel does not result necessarily mean to benefit from the compute resources (cpu and memory) of more than one node.
Even if ParaView distributes the data and work over the compute nodes this might not happen equally.
Two main issues must be considered:
- Parallel Data Management - the data must be distributed equally across parallel processes to take advantage of resources
- Parallel Work Management - the work must be distributed equally across parallel processes to take advantage of resources
You have less influence on the 'Parallel Work Management', therefore we only discuss 'Parallel Data Management' for now.
Parallel Data Management
Data must be distributed across parallel processes to take advantage of resources.
This distribution can be accomplished by the reader or the D3 filter afterwards.
- fully parallel readers
- Explicit parallel formats use separate files for partitions (.pvti, global.silo)
- Implicit parallel formats – parallel processes figure out what they need (.vti, brick-f-values)
- serial readers + included distribute of data
- first process needs enough memory for entire dataset plus additional space for partitioning
- fully serial readers
- all data will end up in first ParaView process
- you need to distribute data manually using D3 filter
- attention: D3 filter outputs unstructured grid, which might need more memory
More about parallel file formats can be found here.
Test Parallel Data Mangement
- Load your data
- Add filter 'Process Id Scalars'
- Segments are colored by which process handles them
How to speed-up ParaView even more
- https://blog.kitware.com/accelerated-filters-in-paraview-5/
- http://www.paraview.org/Wiki/StreamingParaView
- http://www.paraview.org/ParaView/Doc/Nightly/www/py-doc/paraview.simple.TemporalCache.html
In-Situ Visulalization with ParaView Catalyst

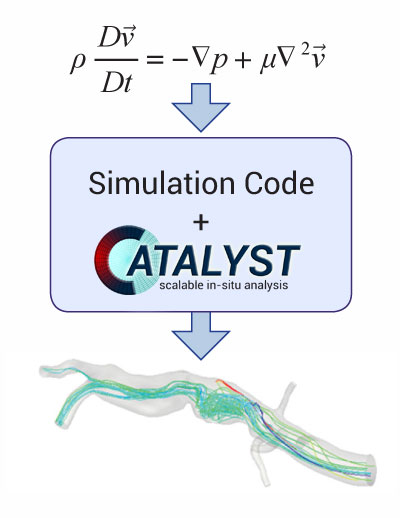 ParaView provides a library for in-situ visualization and analysis that is called Catalyst.
ParaView provides a library for in-situ visualization and analysis that is called Catalyst.
"Catalyst is a light-weight version of the ParaView server library that is designed to be directly embedded into parallel simulation codes to perform in situ analysis at run time." link
Ok, what do you have to do:
- check this webpage html
- instrument your code
- link your code with Catalyst
- start simulation
- connect with GUI to simulation
(more detailed infos will be added - in the mean time check this link)
General Links
ParaView Homepage: http://www.paraview.org
ParaView Wiki: http://www.paraview.org/Wiki/ParaView
ParaView Gallery: http://www.paraview.org/gallery
ParaView for Scientific Data with Python: http://conference.scipy.org/proceedings/scipy2015/pdfs/cory_quammen.pdf
Catalyst Links
Catalyst: http://www.paraview.org/in-situ
Catalyst Wiki: http://www.paraview.org/Wiki/ParaView/Catalyst/Overview
Immersive ParaView
Overview: http://www.paraview.org/immersive/
ParaView Wiki: http://www.paraview.org/Wiki/Immersive_ParaView
Immersive ParaView at JSC: Immersive ParaView on PiCasso Projection System (zam069)
Misc Links
General Cluster: run ParaView on a general cluster
JUVIS: A sample ParaView session with remote rendering
JUVIS: A sample ParaView session with local rendering
any feedback welcomed - j.goebbert@…, h.zilken@…
Attachments (4)
- hardwareComponents.png (203.6 KB ) - added by 8 years ago.
- srun-pvserver-vis.sh (2.2 KB ) - added by 7 years ago.
- 4_1375173693.jpg (27.5 KB ) - added by 6 years ago.
- CatalystFullWorkFlow.png (218.8 KB ) - added by 6 years ago.
Download all attachments as: .zip

